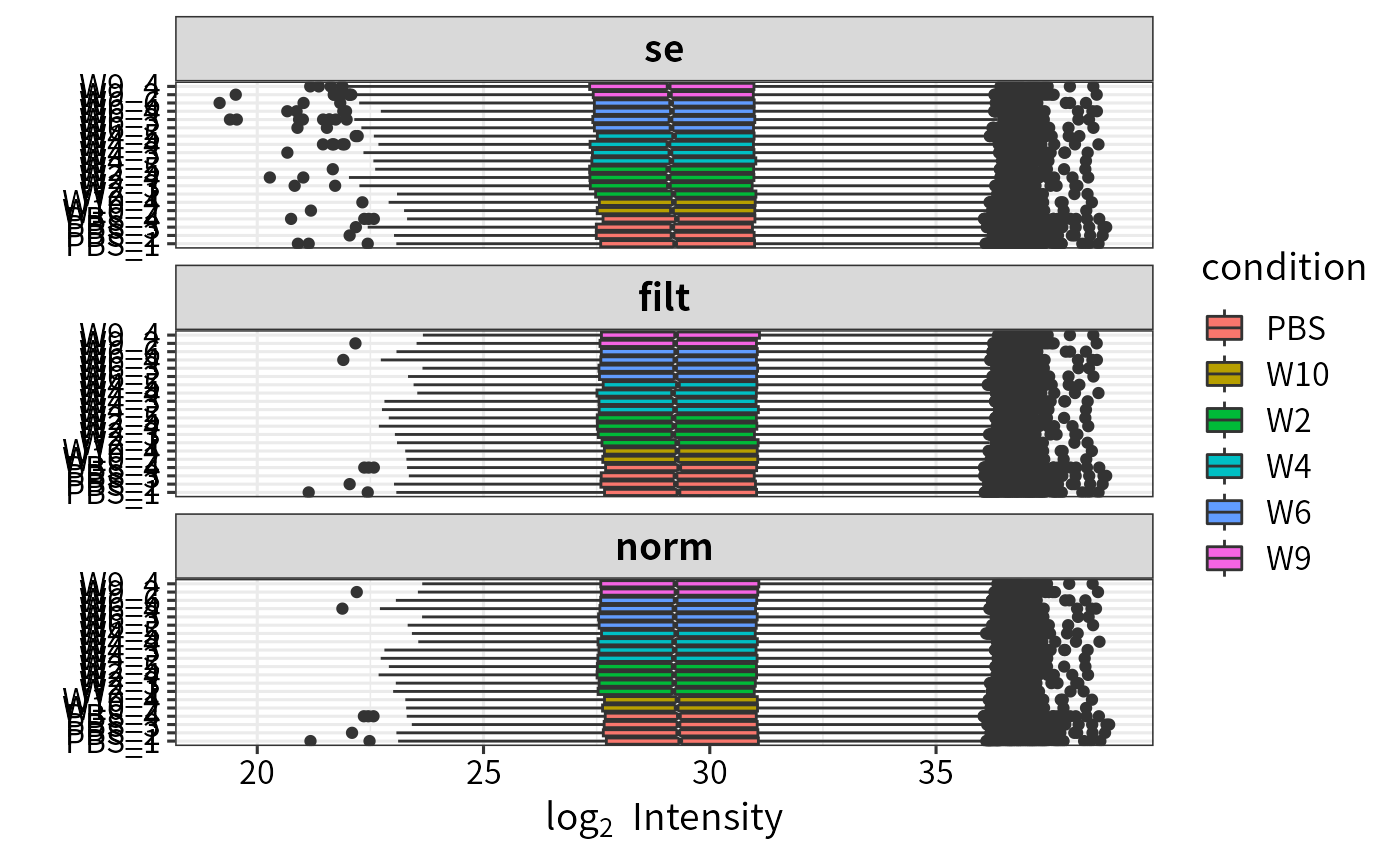
Visualize normalization
plot_normalization.Rdplot_normalization generates boxplots
of all conditions for input objects, e.g. before and after normalization.
Arguments
- se
SummarizedExperiment, Data object, e.g. before normalization (output from
make_se()ormake_se_parse()).- ...
Additional SummarizedExperiment object(s), E.g. data object after normalization (output from
normalize_vsn).
Value
Boxplots of all conditions
for input objects, e.g. before and after normalization
(generated by ggplot).
Adding components and other plot adjustments can be easily done
using the ggplot2 syntax (i.e. using '+')
Examples
# Load example
data(Silicosis_pg)
data <- Silicosis_pg
data_unique <- make_unique(data, "Gene.names", "Protein.IDs", delim = ";")
# Construct SE
ecols <- grep("LFQ.", colnames(data_unique))
se <- make_se_parse(data_unique, ecols,mode = "delim")
# Filter and normalization
filt <- filter_se(se, thr = 0, fraction = 0.4, filter_formula = ~ Reverse != "+" & Potential.contaminant!="+")
#> filter base on missing number is <= 0 in at least one condition.
#> filter base on missing number fraction < 0.4 in each row
#> filter base on giving formula
norm <- normalize_vsn(filt)
#> vsn2: 8762 x 20 matrix (1 stratum).
#> Please use 'meanSdPlot' to verify the fit.
# Plot normalization
plot_normalization(se, filt, norm)