Volcano plot
plot_volcano.Rdplot_volcano generates a volcano plot for a specified contrast.
Usage
plot_volcano(
object,
contrast = get_contrast(object)[1],
add_names = TRUE,
dot_size = 2,
label_size = 3,
label_number = 10,
label_trend = c("all", "up", "down", "none"),
up_color = "#B2182B",
down_color = "#2166AC",
stable_color = "#BEBEBE",
chooseTolabel = NULL,
adjusted = FALSE,
plot = TRUE,
x_symmetry = TRUE,
add_threshold_line = c("none", "intersect", "curve"),
fcCutoff = 1,
pCutoff = 0.05,
curvature = 0.6,
x0_fold = 2,
breaks = NULL,
highlight_PGs_with_few_peptides = F,
peptide_1_color = "#A020F0",
peptide_2_color = "#0000FF",
peptide_column = "Peptides"
)Arguments
- object
SummarizedExperiment, Data object for which differentially enriched proteins are annotated (output from
test_diff()ortest_diff_deg()andadd_rejections()).- contrast
Character(1), Specifies the contrast to plot.
- add_names
Logical(1), Whether or not to plot names.
- dot_size
numeric(1), the size of points.
- label_size
numeric(1), Sets the size of name labels.
- label_number
Integer(1). The number of plot name. Defalut NULL, label all significant names.
- label_trend
Character(1). one of c("all","up","down"), plot all significant, up-regulated ones or down-regulated ones.
- up_color
The color of upregulated points
- down_color
The color of downregulated points
- stable_color
The color of stable points
- chooseTolabel
Character, Specify names to plot, no matter whether they are significant.
- adjusted
Logical(1), Whether or not to use adjusted p values.
- plot
Logical(1), If
TRUE(default) the volcano plot is produced. Otherwise (ifFALSE), the data which the volcano plot is based on are returned.- x_symmetry
Logical(1), if The X-axis is symmetric based on the 0.
- add_threshold_line
Character(1), one of c("none", "intersect", "curve"). "intersect" draw a intersect cutoff line according fcCutoff and pCutoff. "curve" draw a curve cutoff line according curvature and x0_fold.
- fcCutoff
numeric(1), Cutoff of L2FC.
- pCutoff
numeric(1), Cutoff of p or p adjusted value
- curvature
numeric(1), Curvature of curve cutoff line
- x0_fold
numeric(1), The fold of x0 to Sigma.
- breaks
Numeric vector, the position of x-axis label. eg: seq(-10,10,by = 5), c(-2, -1, 0, 1, 2).
- highlight_PGs_with_few_peptides
logic(1), mark the proteins with few peptides by
peptide_1_colorandpeptide_2_color. Require the peptide number variable(specified bypeptide_column) in object.- peptide_1_color
The color of points with 1 peptide.
- peptide_2_color
The color of points with 2 peptide.
- peptide_column
Character(1), the varable column name storing the peptide number.
Value
A volcano plot (generated by ggplot)
Examples
# Load example
data(Silicosis_pg)
data <- Silicosis_pg
data_unique <- make_unique(data, "Gene.names", "Protein.IDs", delim = ";")
# Differential test
ecols <- grep("LFQ.", colnames(data_unique))
se <- make_se_parse(data_unique, ecols,mode = "delim")
filt <- filter_se(se, thr = 0, fraction = 0.4, filter_formula = ~ Reverse != "+" & Potential.contaminant!="+")
#> filter base on missing number is <= 0 in at least one condition.
#> filter base on missing number fraction < 0.4 in each row
#> filter base on giving formula
norm <- normalize_vsn(filt)
#> vsn2: 8762 x 20 matrix (1 stratum).
#> Please use 'meanSdPlot' to verify the fit.
imputed <- impute(norm, fun = "MinProb", q = 0.05)
#> Imputing along margin 2 (samples/columns).
#> [1] 0.3026531
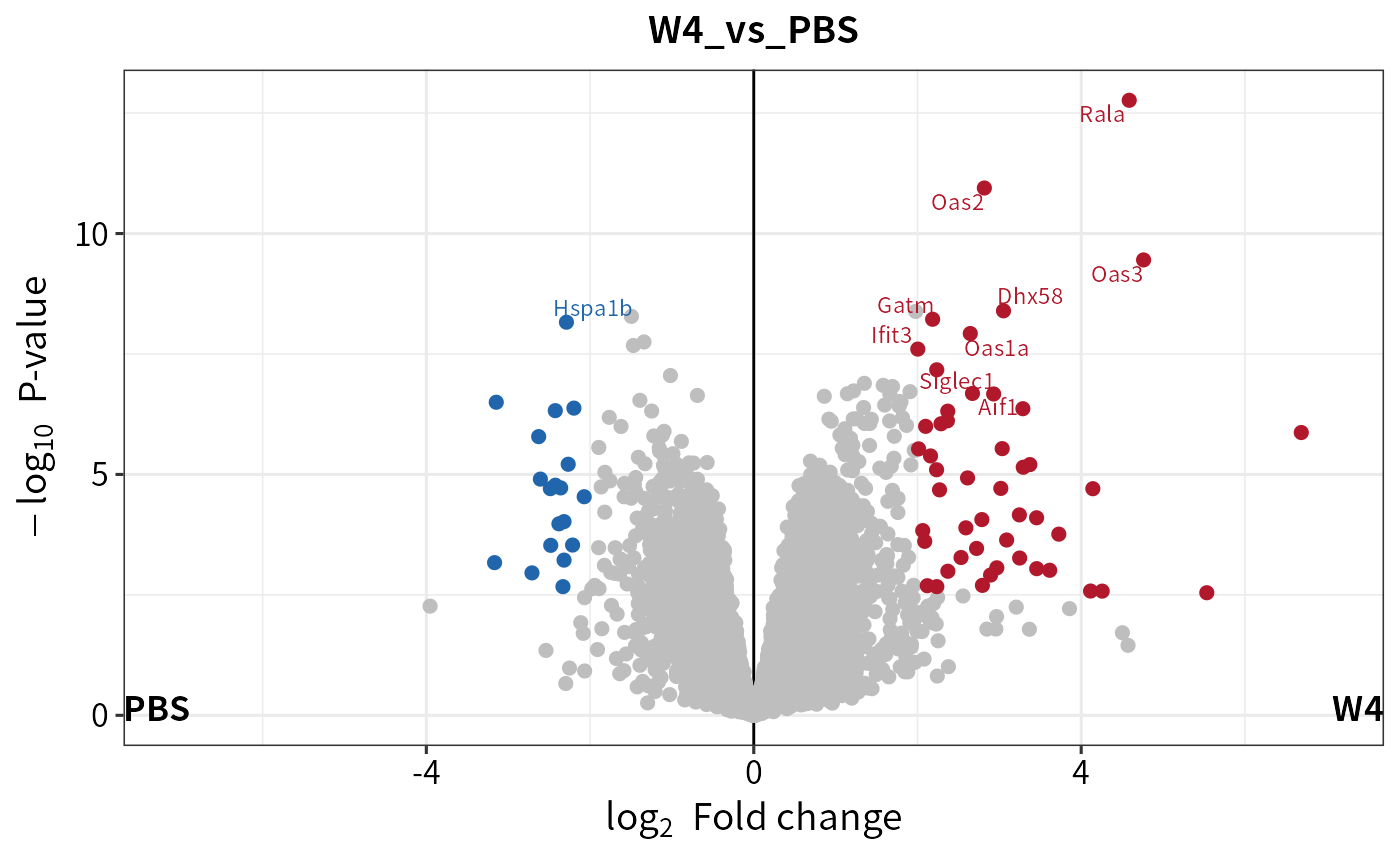
diff <- test_diff(imputed, type = "control", control = c("PBS"), fdr.type = "Storey's qvalue")
#> Tested contrasts: W10_vs_PBS, W2_vs_PBS, W4_vs_PBS, W6_vs_PBS, W9_vs_PBS
#> Storey's qvalue
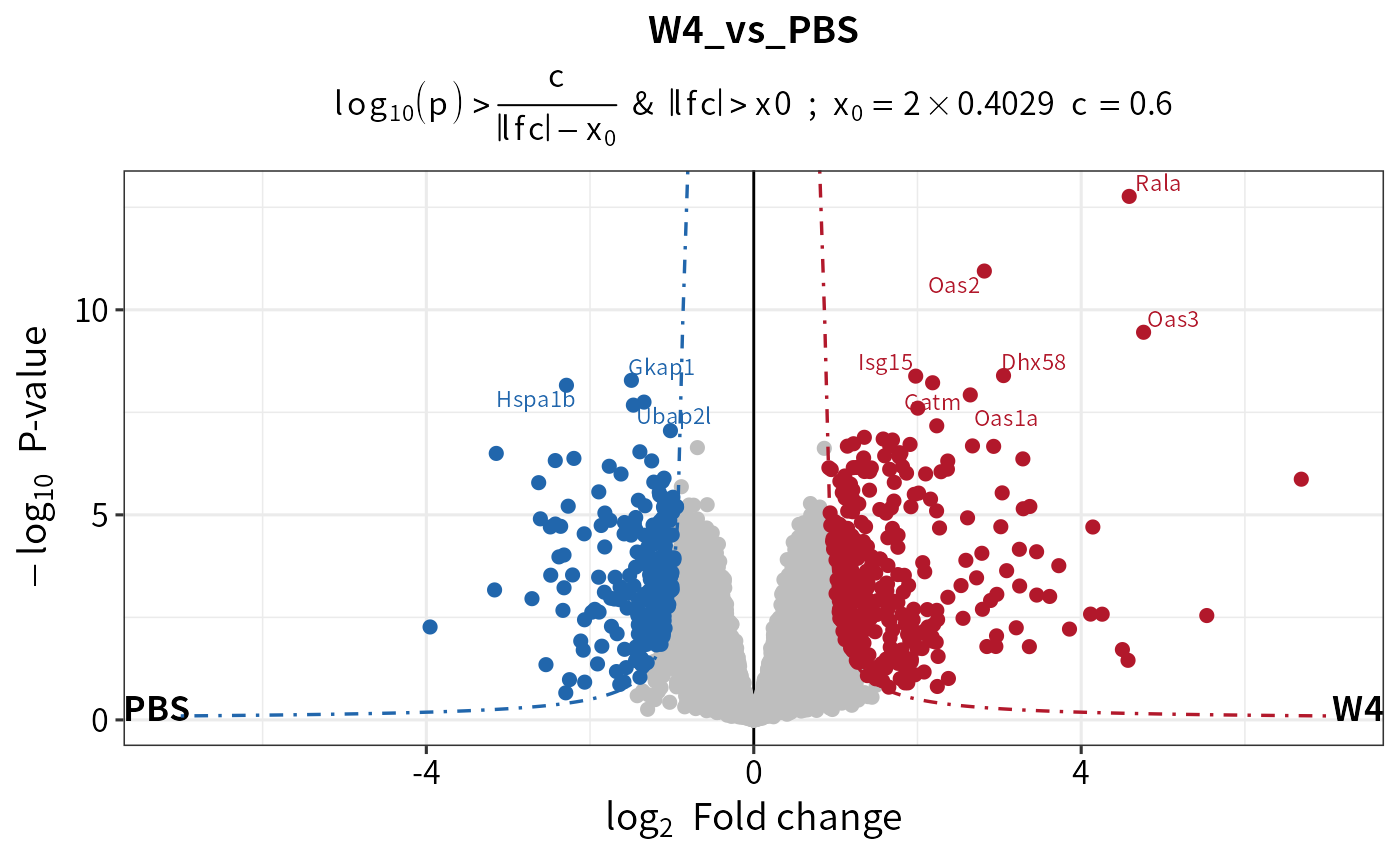
dep <- add_rejections(diff, alpha = 0.01,lfc = 2)
plot_volcano(dep,contrast = "W4_vs_PBS")
 dep <- add_rejections(diff, thresholdmethod = "curve")
plot_volcano(dep,contrast = "W4_vs_PBS",add_threshold_line= "curve")
#> add curve threshold line. Sigma = 0.402890639020025, x0 = 0.80578127804005, curvature = 0.6
dep <- add_rejections(diff, thresholdmethod = "curve")
plot_volcano(dep,contrast = "W4_vs_PBS",add_threshold_line= "curve")
#> add curve threshold line. Sigma = 0.402890639020025, x0 = 0.80578127804005, curvature = 0.6