
Time-course clustering
get_tc_cluster.RdExpression pattern clustering for timecourse or multi-group experiment via
timeclust()
Usage
get_tc_cluster(
x,
ht_mat,
exp_design,
groupby = "condition",
group_order = NULL,
k = 6,
dist = "euclidean",
color = c("RdBu", "RdYlBu", "RdYlGn", "BrBG", "PiYG", "PRGn", "PuOr", "RdGy",
"Spectral"),
col_limit = 4,
row_font_size = 5,
col_font_size = 5,
heatmap_width = 3,
heatmap_height = 5,
seed = NULL
)Arguments
- x
a SummarizedExperiment or DEGdata object.
- ht_mat
Data.frame or matrix, the expresiong assay. If x is provided, be reset to assay(x).
- exp_design
Data.frame, the experiment design. If x is provided, be reset to colData(x)
- groupby
Character(1), group by which column in experiment design.
- group_order
Character vector or factor, the order of groups in the
groupbycolumn.- k
Integer(1), the number of clusters.
- color
Character(1), sets the color panel (from RColorBrewer). It can be a palette name in "RdBu", "RdYlBu", "RdYlGn", "BrBG", "PiYG", "PRGn", "PuOr", "RdGy", "Spectral".
- col_limit
Numeric(1), sets the outer limits of the color scale.
- row_font_size
Integer(1), font size of row name
- col_font_size
Integer(1), font size of columns label.
- heatmap_width
Numeric(1)
- heatmap_height
Numeric(1)
- seed
Integer(1) or NULL, the random seed.
Value
A list contains:
a cluster heatmap(Heatmap object from ComplexHeatmap),
the cluster result for heatmap(data.frame),
the output of
timeclustand the seed.
Examples
# Load sample
data(Silicosis_pg)
data <- Silicosis_pg
data_unique <- make_unique(data, "Gene.names", "Protein.IDs", delim = ";")
# Make SummarizedExperiment
ecols <- grep("LFQ.", colnames(data_unique))
se <- make_se_parse(data_unique, ecols, mode = "delim", sep = "_")
filt <- filter_se(se, thr = 0, fraction = 0.3, filter_formula = ~ Reverse != "+" & Potential.contaminant!="+")
#> filter base on missing number is <= 0 in at least one condition.
#> filter base on missing number fraction < 0.3 in each row
#> filter base on giving formula
norm <- normalize_vsn(filt)
#> vsn2: 8832 x 20 matrix (1 stratum).
#> Please use 'meanSdPlot' to verify the fit.
imputed <- impute(norm, fun = "MinProb", q = 0.05)
#> Imputing along margin 2 (samples/columns).
#> [1] 0.3026538
diff <- test_diff(imputed,type = "control", control = "PBS")
#> Tested contrasts: W10_vs_PBS, W2_vs_PBS, W4_vs_PBS, W6_vs_PBS, W9_vs_PBS
#> Strimmer's qvalue(t)
dep <- add_rejections(diff)
# Expression pattern cluster
TC <- get_tc_cluster(get_signicant(dep),
group_order = c("PBS","W2","W4","W6","W9","W10"), # set the group order
heatmap_width = 2.5, heatmap_height = 5
)
#> aabb[1] "start timeclust"
#> beging 'cm' cluster by dist = 'euclidean'
#> [1] "finish heatmap"
TC$ht # Heatmap
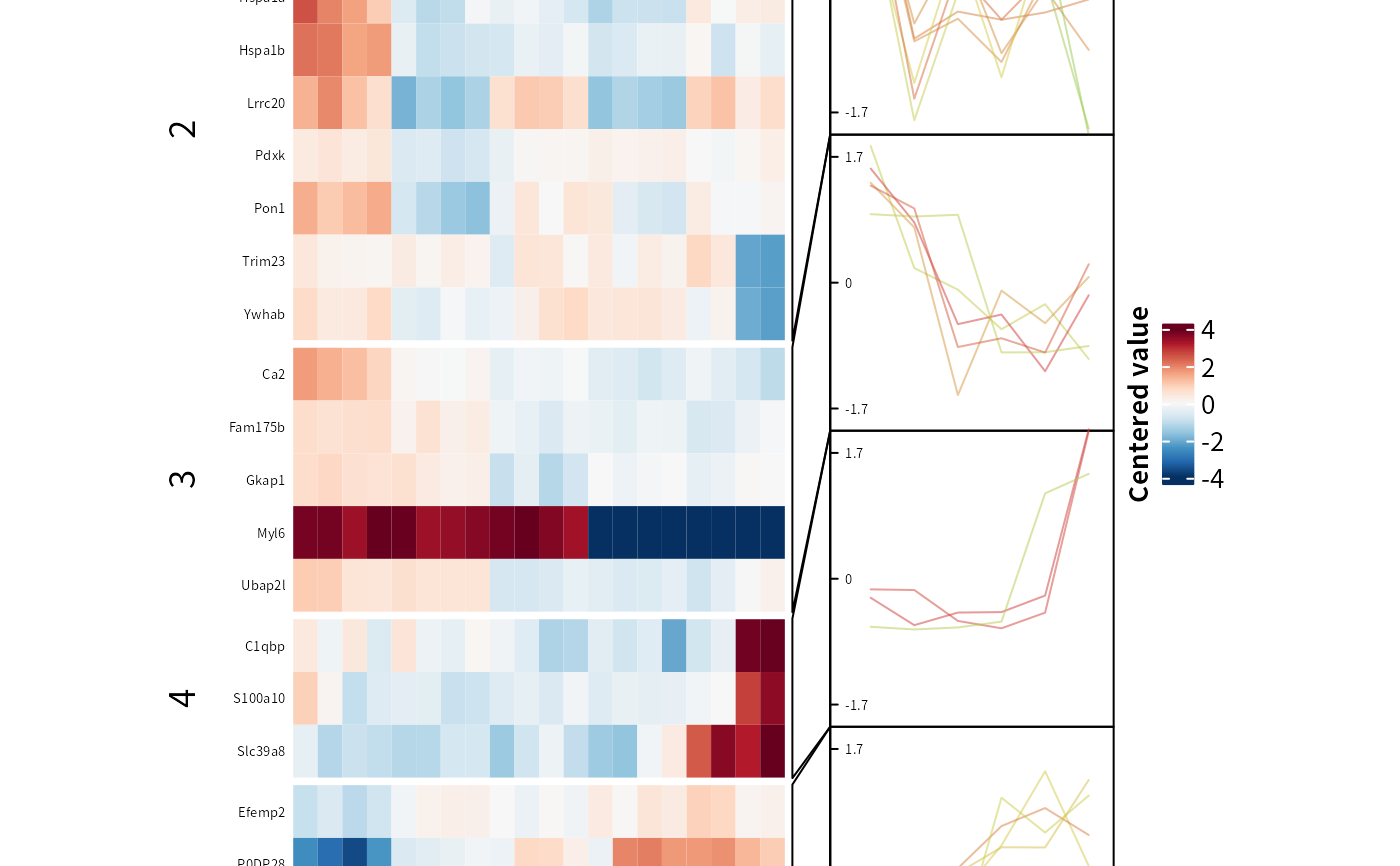 head(TC$res)
#> name Timecourse_cluster mebership PBS_1
#> C1qbp C1qbp 4 0.886304715790769 0.420263091052544
#> Ca2 Ca2 3 0.411242340092725 1.69065263119242
#> Cldn18 Cldn18 2 0.686617687623295 1.23958646631143
#> Dhx58 Dhx58 1 0.846778402740017 -2.06544095513807
#> Dnmt3a Dnmt3a 6 0.552528926838831 -0.904896251325489
#> Efemp2 Efemp2 5 0.505110922852042 -0.930272399356138
#> PBS_2 PBS_3 PBS_4
#> C1qbp -0.195529409711714 0.424599288402426 -0.575735101668087
#> Ca2 1.41759313041917 1.21867872572312 0.871398543111233
#> Cldn18 1.14834276230686 1.17683392038859 0.649627636229976
#> Dhx58 -1.96084347617414 -2.93920556318203 -3.1943284449183
#> Dnmt3a -0.939565872453979 -1.17776444420126 -0.906231190648125
#> Efemp2 -0.605958902335448 -1.08561736048544 -0.812065013310434
#> W2_1 W2_3 W2_4
#> C1qbp 0.515549989532623 -0.204071405680601 -0.36550912598274
#> Ca2 0.098613225762044 0.0159171430763649 -0.0158498148229711
#> Cldn18 -0.360596970057859 -0.47187623920922 -0.4255120562602
#> Dhx58 0.746457200193323 1.24456979871951 0.821772070624508
#> Dnmt3a 0.783357254120432 1.08808001389562 0.868693353844201
#> Efemp2 -0.15358956170402 0.174213060034393 0.249851981034421
#> W2_5 W4_2 W4_3
#> C1qbp 0.062132795471836 -0.162074321207182 -0.485304611494996
#> Ca2 0.100742763687713 -0.37424848888098 -0.119820470177196
#> Cldn18 -0.288477991839169 0.330153178862002 0.239899871574369
#> Dhx58 1.03333640641943 1.13127573675511 0.192532251407595
#> Dnmt3a 0.749978466197163 0.583297318826038 -0.395325255498001
#> Efemp2 0.214905350869383 0.00808615857910056 -0.244696885643624
#> W4_4 W4_5 W6_2
#> C1qbp -1.24162814651021 -1.16854438888593 -0.443699676074125
#> Ca2 -0.181299577487948 -0.0116517269610839 -0.460001709658762
#> Cldn18 0.199424707360595 0.567415080217049 -0.619790712033538
#> Dhx58 0.716047237780469 0.00185594051893645 0.123212926214716
#> Dnmt3a 0.277692440227106 -0.312137949657195 -0.0517147839181824
#> Efemp2 0.0411470168599486 -0.173965660789197 0.363980847906301
#> W6_3 W6_4 W6_6
#> C1qbp -0.792023214384841 -0.480044206757867 -2.07162607062625
#> Ca2 -0.530563532665759 -0.776238614295806 -0.539418285530573
#> Cldn18 -0.783321687943666 -0.794706626067534 -0.405821220765933
#> Dhx58 0.734945929150886 0.642967460601366 0.555262089321882
#> Dnmt3a 0.477148540207022 0.228572037733063 0.393591813383914
#> Efemp2 0.0413562665920004 0.50074823953592 0.342584869825163
#> W9_2 W9_4 W10_2
#> C1qbp -0.77498614221431 -0.342256735421753 3.8874092603213
#> Ca2 -0.163821633610759 -0.476498689126373 -0.713390780262202
#> Cldn18 -0.129757499196334 -0.0287980472910228 -0.414560565381205
#> Dhx58 0.393764307310281 0.481596331711717 0.840155068871187
#> Dnmt3a -0.439023661513279 -0.22878860702064 -0.0366646751473709
#> Efemp2 0.925971985604367 0.835542242088724 0.115609350582677
#> W10_4
#> C1qbp 3.99307813183992
#> Ca2 -1.05079283949163
#> Cldn18 -0.828064007205182
#> Dhx58 0.500067683811611
#> Dnmt3a -0.0582985470510273
#> Efemp2 0.192168414111929
head(TC$res)
#> name Timecourse_cluster mebership PBS_1
#> C1qbp C1qbp 4 0.886304715790769 0.420263091052544
#> Ca2 Ca2 3 0.411242340092725 1.69065263119242
#> Cldn18 Cldn18 2 0.686617687623295 1.23958646631143
#> Dhx58 Dhx58 1 0.846778402740017 -2.06544095513807
#> Dnmt3a Dnmt3a 6 0.552528926838831 -0.904896251325489
#> Efemp2 Efemp2 5 0.505110922852042 -0.930272399356138
#> PBS_2 PBS_3 PBS_4
#> C1qbp -0.195529409711714 0.424599288402426 -0.575735101668087
#> Ca2 1.41759313041917 1.21867872572312 0.871398543111233
#> Cldn18 1.14834276230686 1.17683392038859 0.649627636229976
#> Dhx58 -1.96084347617414 -2.93920556318203 -3.1943284449183
#> Dnmt3a -0.939565872453979 -1.17776444420126 -0.906231190648125
#> Efemp2 -0.605958902335448 -1.08561736048544 -0.812065013310434
#> W2_1 W2_3 W2_4
#> C1qbp 0.515549989532623 -0.204071405680601 -0.36550912598274
#> Ca2 0.098613225762044 0.0159171430763649 -0.0158498148229711
#> Cldn18 -0.360596970057859 -0.47187623920922 -0.4255120562602
#> Dhx58 0.746457200193323 1.24456979871951 0.821772070624508
#> Dnmt3a 0.783357254120432 1.08808001389562 0.868693353844201
#> Efemp2 -0.15358956170402 0.174213060034393 0.249851981034421
#> W2_5 W4_2 W4_3
#> C1qbp 0.062132795471836 -0.162074321207182 -0.485304611494996
#> Ca2 0.100742763687713 -0.37424848888098 -0.119820470177196
#> Cldn18 -0.288477991839169 0.330153178862002 0.239899871574369
#> Dhx58 1.03333640641943 1.13127573675511 0.192532251407595
#> Dnmt3a 0.749978466197163 0.583297318826038 -0.395325255498001
#> Efemp2 0.214905350869383 0.00808615857910056 -0.244696885643624
#> W4_4 W4_5 W6_2
#> C1qbp -1.24162814651021 -1.16854438888593 -0.443699676074125
#> Ca2 -0.181299577487948 -0.0116517269610839 -0.460001709658762
#> Cldn18 0.199424707360595 0.567415080217049 -0.619790712033538
#> Dhx58 0.716047237780469 0.00185594051893645 0.123212926214716
#> Dnmt3a 0.277692440227106 -0.312137949657195 -0.0517147839181824
#> Efemp2 0.0411470168599486 -0.173965660789197 0.363980847906301
#> W6_3 W6_4 W6_6
#> C1qbp -0.792023214384841 -0.480044206757867 -2.07162607062625
#> Ca2 -0.530563532665759 -0.776238614295806 -0.539418285530573
#> Cldn18 -0.783321687943666 -0.794706626067534 -0.405821220765933
#> Dhx58 0.734945929150886 0.642967460601366 0.555262089321882
#> Dnmt3a 0.477148540207022 0.228572037733063 0.393591813383914
#> Efemp2 0.0413562665920004 0.50074823953592 0.342584869825163
#> W9_2 W9_4 W10_2
#> C1qbp -0.77498614221431 -0.342256735421753 3.8874092603213
#> Ca2 -0.163821633610759 -0.476498689126373 -0.713390780262202
#> Cldn18 -0.129757499196334 -0.0287980472910228 -0.414560565381205
#> Dhx58 0.393764307310281 0.481596331711717 0.840155068871187
#> Dnmt3a -0.439023661513279 -0.22878860702064 -0.0366646751473709
#> Efemp2 0.925971985604367 0.835542242088724 0.115609350582677
#> W10_4
#> C1qbp 3.99307813183992
#> Ca2 -1.05079283949163
#> Cldn18 -0.828064007205182
#> Dhx58 0.500067683811611
#> Dnmt3a -0.0582985470510273
#> Efemp2 0.192168414111929